Fantastic news: Our miCube microscopy platform and the recent publication “Visualisation of dCas9 target search in vivo using an open-microscopy framework” [link] was featured in Nature Methods [link]. Thank you Dr. Strack!
Category: miCube
Publication: Visualisation of dCas9 target search in vivo using an open-microscopy framework
K.J.A. Martens, S. van Beljouw, S. van der Els, J.N.A. Vink, S. Baas, G.A. Vogelaar, S.J.J. Brouns, P. van Baarlen, M. Kleerebezem, J. Hohlbein, Nature Communications, 10, 3552, 2019, [link]
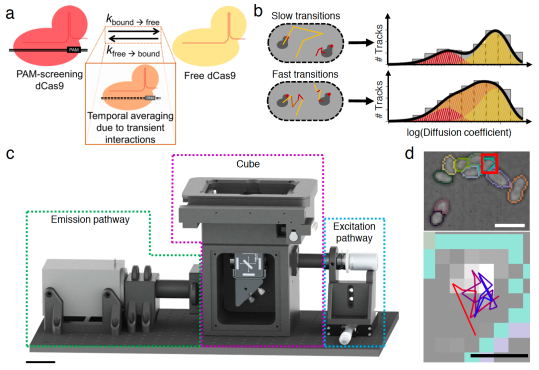
CRISPR-Cas9 is widely used in genomic editing, but the kinetics of target search and its relation to the cellular concentration of Cas9 have remained elusive. Effective target search requires constant screening of the protospacer adjacent motif (PAM) and a 30 ms upper limit for screening was recently found. To further quantify the rapid switching between DNA-bound and freely-diffusing states of dCas9, we developed an open-microscopy framework, the miCube, and introduce Monte-Carlo diffusion distribution analysis (MC-DDA). Our analysis reveals that dCas9 is screening PAMs 40% of the time in Gram-positive Lactoccous lactis, averaging 17 ± 4 ms per binding event. Using heterogeneous dCas9 expression, we determine the number of cellular target-containing plasmids and derive the copy number dependent Cas9 cleavage. Furthermore, we show that dCas9 is not irreversibly bound to target sites but can still interfere with plasmid replication. Taken together, our quantitative data facilitates further optimization of the CRISPR-Cas toolbox.
News: The miCube moved to Github
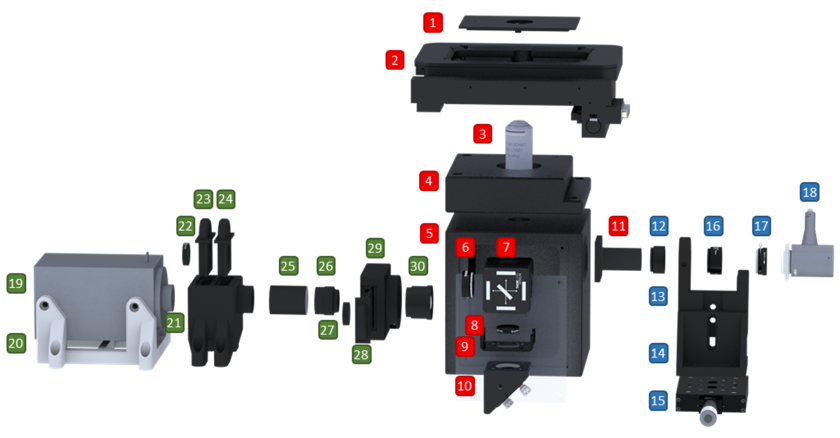
Our open (single-molecule) microscopy project, the #miCube, is now on Github [link]! The page shows a detailed and updated overview of the components, some information on phasor-based SMLM, and many links to similar open hardware projects.
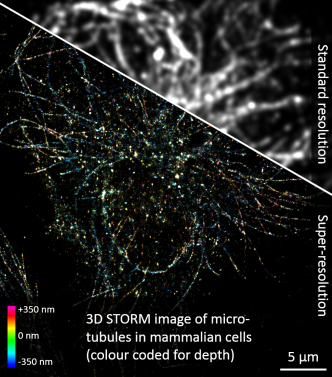
miCube: 3D super-resolution microscopy
Here the specs: 50k frames, 850k localisations, 6 min for data analysis. Many thanks to Sander Baas, Koen Martens and Arjen Bader, with cells provided by Gert-Jan Bakker and Ben Joosten (both Radboudumc, Nijmegen).
News: (Update) miCube: Rethinking modular fluorescence microscopy
Hurrah, we finalised the design of our miCube (V0.1)! Details can be found [here] and include part numbers, CAD-drawings and STL files for CNC machining/milling or 3D printing published under a Creative Common license. Big shout out to Sander Baas and Koen Martens. If you have any questions, remarks or ideas, please feel to contact me.

miCube V0.1: first tests
We finally managed to perform some test experiments and it is looking very promising! We tracked small fluorescent quantum dots (< 5 nm diameter) diffusing in water (20% glycerol) and we were happy to learn that even for such a demanding application new sCMOS-based cameras are doing just fine. Here a picture of the current assembly. More soon!
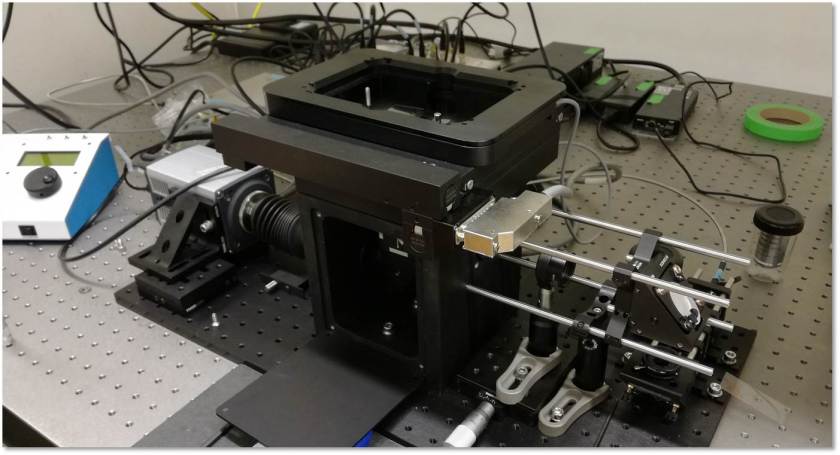
News: miCube V0.1 prototyping
We just got the very first version from the workshop to test the general design layout. Looks promising. The cube went back to drill and tap a few more holes and to receive a black anodisation.

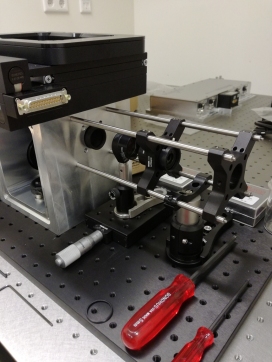
Introducing miCube V0.1: Rethinking modular fluorescence microscopy
Update: Follow the link or Twitter (#miCube) for the lastest information.
Fluorescence microscopy is an extremely powerful and versatile technique contributing to many areas of the life sciences. Especially variants featuring the ability to monitor single-molecule fluorescence, however, require sophisticated instrumentation that is either very expensive when bought commercially (>> 100 kEuro) or demands extensive expertise in optics and engineering.
Here we present an open and modular hardware framework aiming for
- cost effectiveness: build your own starting at 20k Euro (~100 kEuro for state of the art capabilities)
- modularity: all parts can be accessed and replaced by the user
- simplicity: set up the microscope in a few hours without prior knowledge
- customizability: confocal or widefield/TIRF microscopy,…
- openness: part lists and drawings will be made available
- stability and throughput: minimizing drift and utilise well plate scanners
Interested? Drop me a line. We are currently working together with several academic labs to bring their ideas to life and develop the miCube concept further.

For a similar concept, please also visit http://wosmic.org/.