For the start of the academic year, we welcome new lab members: Erwin Dijkstra started with his MSc thesis on spectrally resolved super-resolution imaging, Victor Pools will help us as a research assistant with cloning and single-molecule particle tracking, and Konstantin Speckner is taking a short break from his PhD in Bayreuth to learn more about single-molecule and single-cell techniques. Welcome on board!
Category: news
News: Presentation on open microscopy and in-vivo single-molecule CRISPR-Cas
In January 2021, I had the pleasure to present some of our recent work as part of the Imaging ONEWORLD series initiated by the Royal Society of Microscopy. Topics I talked about include: open-source microscopy (#miCube), accelerated single-molecule localisation analysis (#SMLM) using phasor analysis, diffusion distribution analysis (#anaDDA), and in vivo single-particle tracking of CRISPR-Cas9.
News: NWO Take-off (phase I) granted & new lab members
Happy to share the news that we received a NWO Take-off (phase 1) grant for “Spectrally-resolved single-molecule localization microscopy to go”. Looking forward working with Niels and the people from Cairn Research Ltd, Confocal.nl, Wageningen University & Research on this exciting project to bring more colours into microscopy.
We further say good bye to Mattia, who will continue working with my new colleague Dr. Sonja Schmid (see her new and shiny webpage here). Hope we can get all those great work of yours published soon!
We also welcome two new BSc students, Mink Neeleman and Casper Peters, who will work on CRISPR-Cas, live cell imaging and microfluidics. Welcome on board!
News: Goodbye Koen and welcome Cleo, Martijn, Niels and Simon
After recently defending his PhD thesis titled “Pinpointing macromolecular motion. Single-particle tracking fluorescence microscopy advances and applications” with distinction (cum laude!), Koen Martens left the group for a postdoc position in the lab of Dr. Ulrike Endesfelder (CMU, Pittsburgh). All the best, Koen, you will be missed!
A big welcome to Martijn Gobes and Cleo Bagchus who both joined the group for their MSc thesis. Martijn will improve our computational workflow for super-resolution microscopy (his first ImageJ plugin for fast temporal median filtering is available here). Cleo will continue our work on single-particle tracking in live cells together with Simon van de Els, who joined the lab as a (semi) post-doctoral research assistant.
Last but not least, we welcome Niels Zijlstra as a guest scientist from the Cordes lab in Munich. Niels will work on combining the miCube platform with spectrally resolved super-resolution imaging.
News: IPM-3 and R2I proposals granted
Good news! Two proposals got granted!
For the first one (IPM-3, Innovation Program Microbiology, an initiative from the Laboratory of Microbiology at Wageningen University and Research), we will use single-particle tracking Photo-Activated Localisation Microscopy (sptPALM) to study CRISPR-Cas in live bacteria. Looking forward collaborating with Raymond Staals from the Laboratory of Microbiology. A one-year post-doc position will be available, please contact me (with CV) for further details.
For the second, the Road-to-Innovation business development grant, we will work on a neat idea for spectrally resolved single-molecule localisation microscopy.
News: Welcome to Mariska Brüls and Dani Kortekaas
With Mariska, the third PhD student in our NWO funded LocalBioFood project on super-resolution based localisation of biomolecules at food-related interfaces has started. Mariska started her PhD thesis in the Voets lab in Eindhoven and will relocate to our lab in early 2022.
Dani started his BSc thesis in the lab during Covid-19 times. He will look into improving our computational workflow for single-particle tracking in live cells.
News: Welcome to Ezra Bekkering
Ezra just started his BSc thesis in a new project together with the Laboratory of Microbiology (Wen Wu & Dr. Raymond Staals). He will work on visualising CRISPR-Cas interactions in live bacteria using sptPALM.
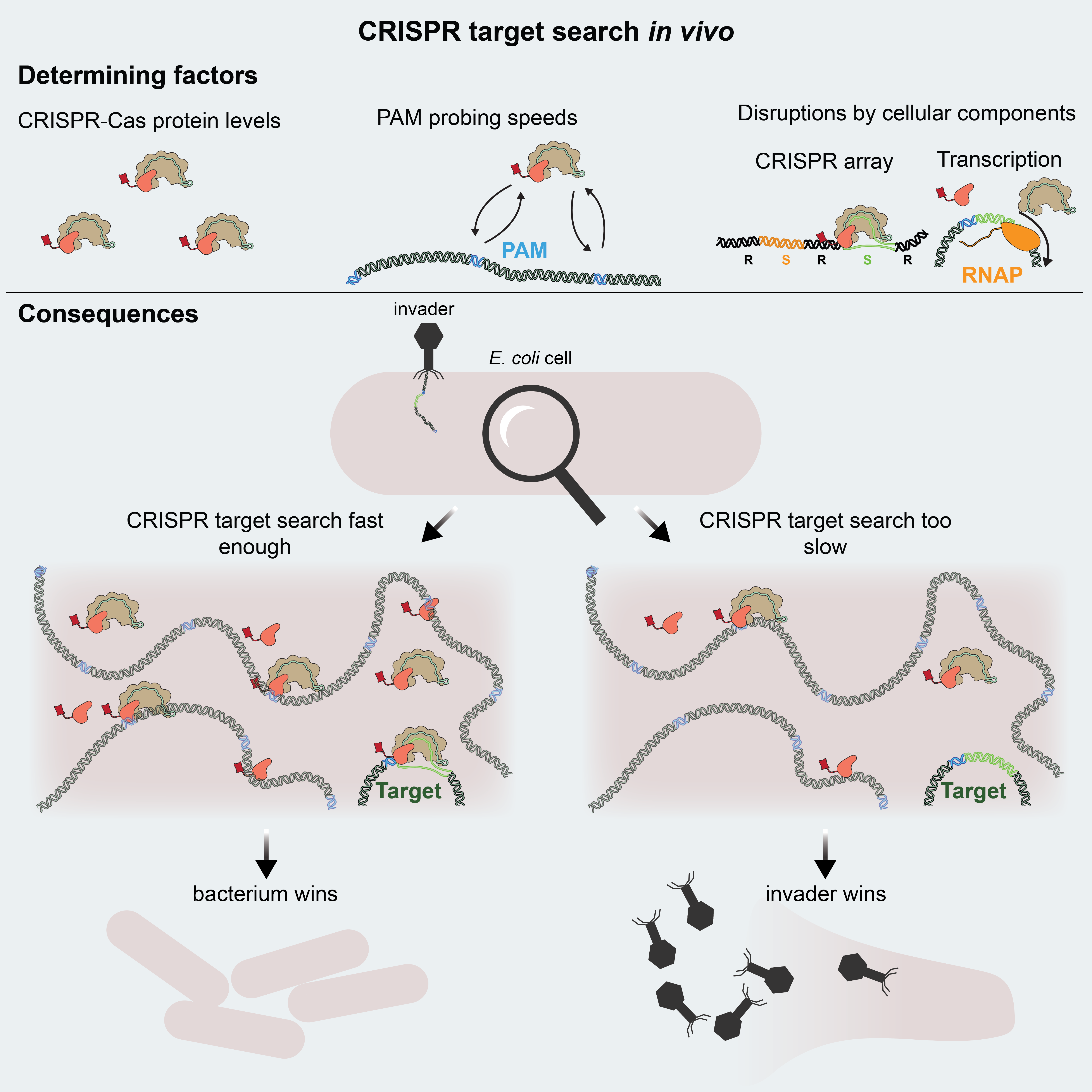
Publication: Direct visualization of native CRISPR target search in live bacteria reveals Cascade DNA surveillance mechanism
CRISPR-Cas systems encode RNA-guided surveil-lance complexes to find and cleave invading DNA elements. While it is thought that invaders are neutralized minutes after cell entry, the mechanism andkinetics of target search and its impact on CRISPRprotection levels have remained unknown. Here, wevisualize individual Cascade complexes in a native type I CRISPR-Cas system. We uncover an exponential relation between Cascade copy number and CRISPR interference levels, pointing to a time-driven arms race between invader replication and target search, in which 20 Cascade complexes provide 50% protection. Driven by PAM-interacting subunitCas8e, Cascade spends half its search time rapidly probing DNA (30 ms) in the nucleoid. We further demonstrate that target DNA transcription and CRISPR arrays affect the integrity of Cascade and affect CRISPR interference. Our work establishes the mechanism of cellular DNA surveillance by Cascade that allows the timely detection of invading DNA in a crowded, DNA-packed environment.
News: miCube featured in Nature Methods
News: Welcome to Šarūnė Ivanovaitė and Elmar van der Wijk
Šarūnė recently started her Erasmus+ internship in the group. She will characterise a variety of fluidic devices that we got our hands on including devices for high throughput smFRET screening, trapping of bacteria and establishing DNA curtains. Elmar started his MSc thesis and will utilise new DNA constructs for smFRET based studies of ARF transcription factor binding.