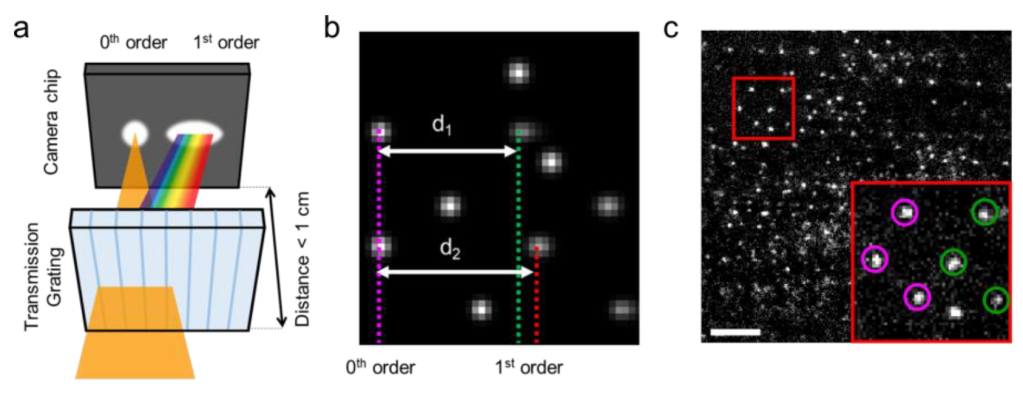
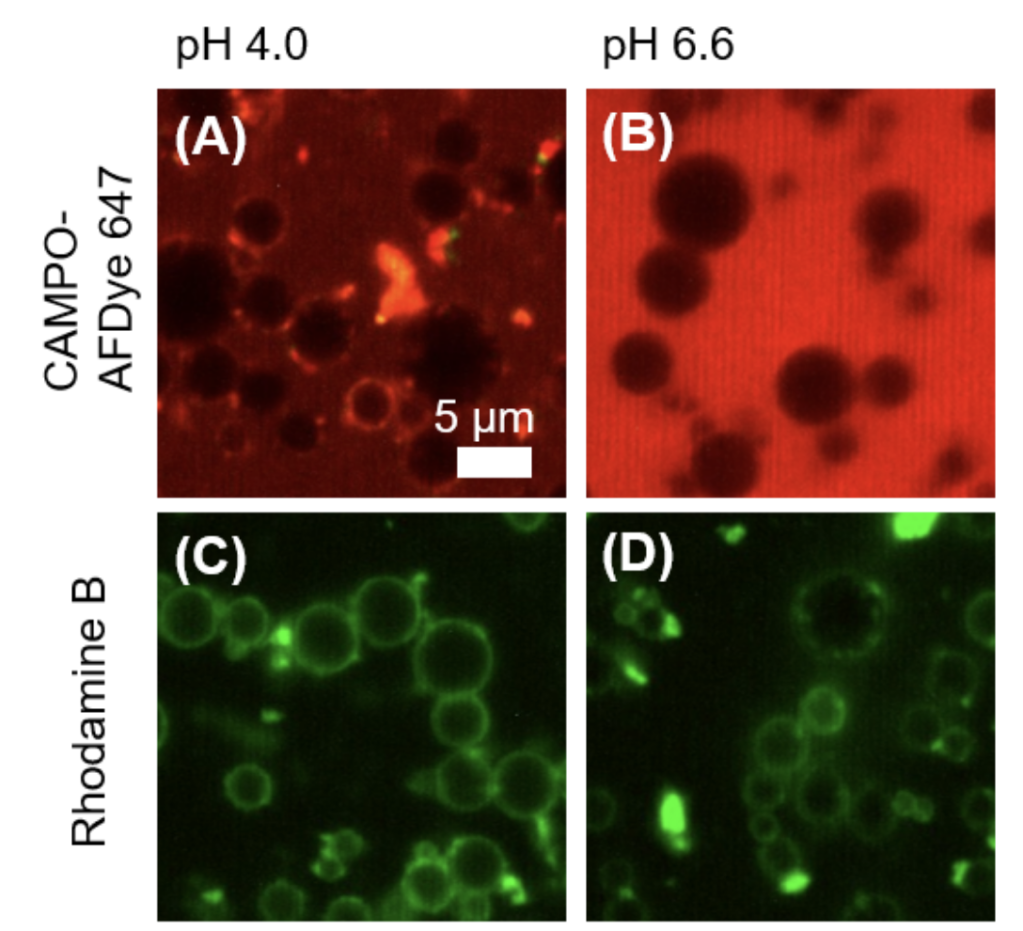
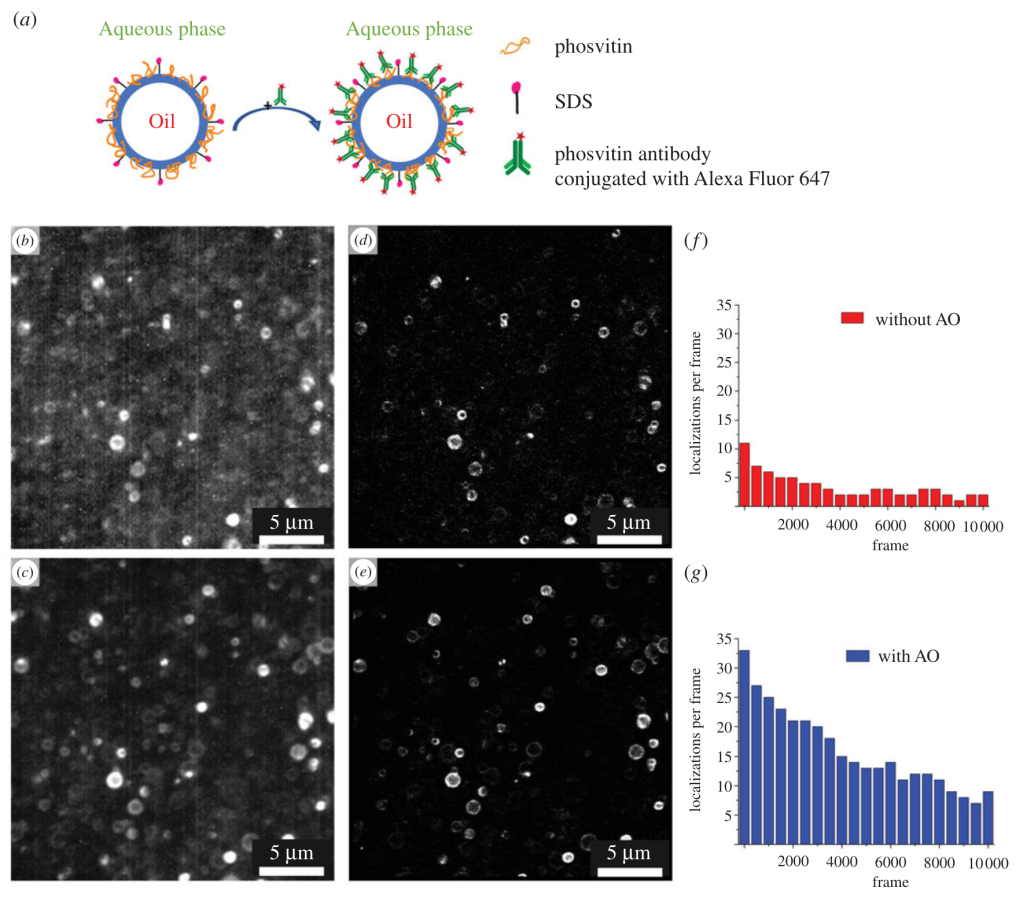
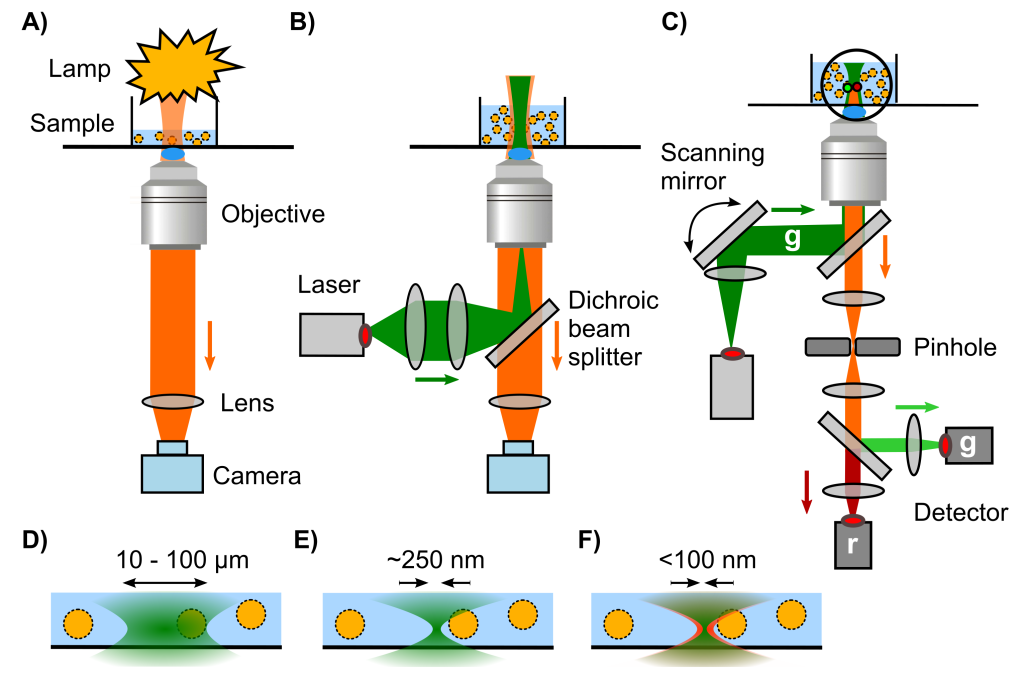
S.Yang, M. Takeuchi, R. Joosten, J. van Duynhoven, H. Friedrich, J. Hohlbein, Food Structure, 40, 100365, 2024, [link], preprint on chemRxiv, [link]
Lipid oxidation is a major cause of product deterioration in protein stabilised oil-in-water food emulsions. The impact of protein emulsifiers on lipid oxidation and the stability depends on the specific type of protein emulsifiers used and the redox conditions in the emulsion. However, the exact impact of these protein emulsifiers at the oil-water interface on lipid oxidation and the mechanism of lipid-protein co-oxidation are currently unknown. Here, we developed a cryo-correlative light and electron microscopy (cryo-CLEM) platform for co-localising the oxidation of lipids and proteins. For this first implementation of cryo-CLEM for food oxidation studies we optimised specifically the part of cryo-fluorescence microscopy (cryo-FM) by adding parts that prevent fogging on the sample and enable homogeneous laser illumination. We showed that lipid oxidation in food emulsions can be observed at cryogenic temperature using fluorescence imaging of the fluorophore BODIPY 665/676 that we employed earlier as a lipid oxidation sensor at room temperature. Using cryo-transmission electron microscopy (cryo-TEM), we observed that more protein aggregates are found at the droplet interfaces in oxidized emulsions compared to fresh emulsions. Our cryo-CLEM platform paves the way for future cryo-correlative oxidation studies of food emulsions.